TECHNOLOGY
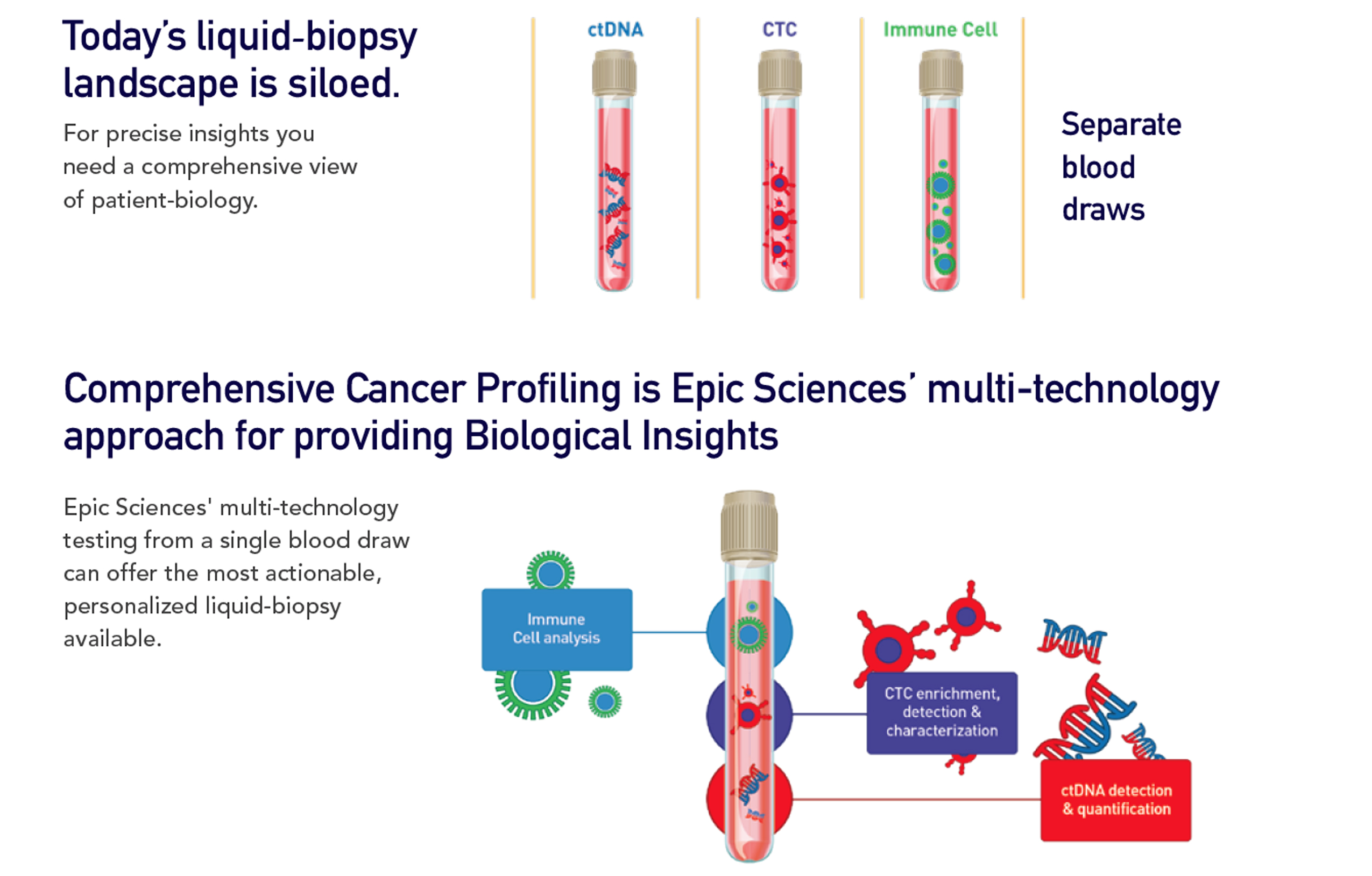
COMPREHENSIVE CANCER PROFILING
Epic Sciences’ revolutionary platform begins with a single, non-invasive blood draw. From this sample, we conduct multiparametric, single-cell analysis of protein expression, cell morphology and single-cell genomics to see the widest biological range of cells with unmatched sensitivity.
Click on a data point to display its corresponding image.
DAPI
CD45
ARV7
CK
Composite
PROCESS: Comprehensive Cancer Profiling provides a unique, interactive 3D model of the cell population, enabling detailed analysis and identification of cells other technologies might miss.
WE SEE WHAT OTHERS DON’T
Epic’s Comprehensive Cancer Profiling approach uses multiple technologies to provide up to three types of analysis all based on a single blood draw. We leverage proven and proprietary CTC capabilities, along with ctDNA and immune-cell analysis. This allows a clearer and more efficient picture for both prostate and breast cancer.
Our enrichment-free approach minimizes the risk of CTCs being missed and reveals both genotypic and phenotypic data. By combining CTC technology with ctDNA and immune-cell analysis, we enable a predictive test that is both minimally invasive and highly accurate.
WATCH OUR COMPREHENSIVE CANCER PROFILING VIDEO
SINGLE-CELL circulating-tumor-cell Analysis
SINGLE-CELL ANALYSIS IS ESSENTIAL TO DETECTING AND UNDERSTANDING CANCER
We can perform single-cell genomic profiling of CTCs and immune-cells and pair the genomics to distinct digital pathology features or protein markers expressed allowing us to see both cell phenotype and genotype.
Comprehensive Cancer Profiling analyzes all nucleated cells within a blood sample at single-cell resolution. Cells from a patient’s blood sample are deposited on glass slides and immunofluorescent staining is used to compare for morphological features, biomarker expression and nuclear integrity. Cell phenotype, biomarker expression, CTCs, ctDNAs, and immune-cells are individually analyzed. Circulating tumor cell DNA is amplified by whole genome amplification (WGA) and analyzed by next-generation sequencing (NGS) for the presence of point mutations, copy number alterations, genome-wide chromosomal instability, ploidy, or genome-wide scarring.
WHOLE GENOME COPY NUMBER VARIATION (CNV)
Epic’s Comprehensive Cancer Profiling can characterize CNV and genomic instability in subclonal populations, and identify specific oncogene amplification or tumor surpressor deletion, genome-wide.
TARGETED RESEQUENCING ASSAYS
Epic’s Comprehensive Cancer Profiling is capable of detecting CTC sub-clonal populations harboring actionable mutations to monitor sensitivity to targeted therapies through the course of treatment. It can also sequence only genes of interest, focusing only on those that are specific drug targets.





